Differential expression analysis with inmoose, the integrated multi-omic open-source environment in Python

We present the differential expression features of InMoose, a Python implementation of R tools limma, edgeR, and DESeq2.
Bridging the gap between R and Python in bulk transcriptomic data analysis with InMoose

We present the differential expression features of InMoose, a Python implementation of R tools limma, edgeR, and DESeq2.
Accelerating antigen-targeting therapy discovery with a scalable pan-cancer bioinformatics platform
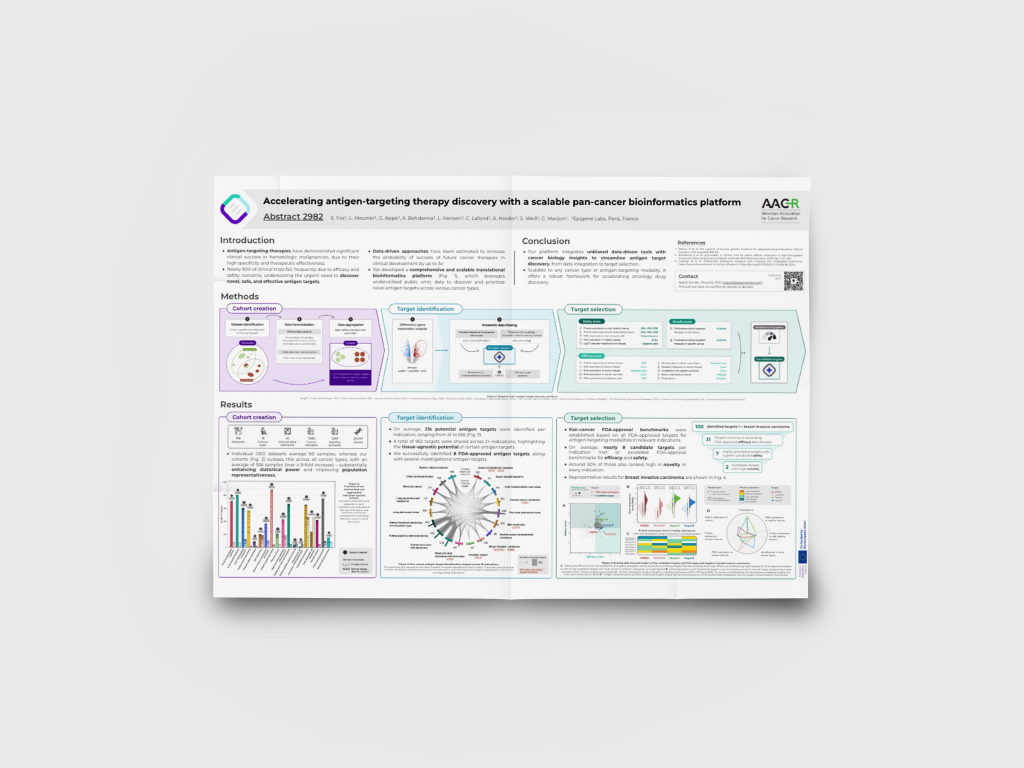
We developed a pan-cancer bioinformatics platform combining patient data with extensive biological and pharmaceutical knowledge.
The evolving landscape of cancer transcriptomics data
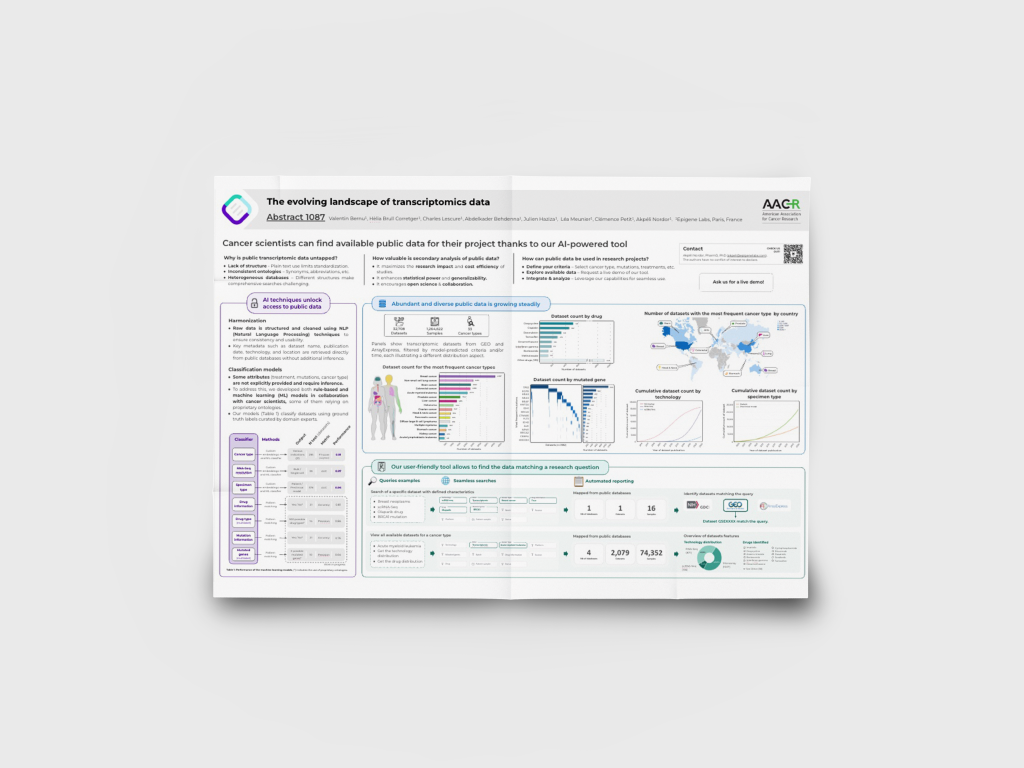
We developed a pan-cancer bioinformatics platform combining patient data with extensive biological and pharmaceutical knowledge.
InMoose: the Integrated Open Source Python Package for Multi-omic Analyses
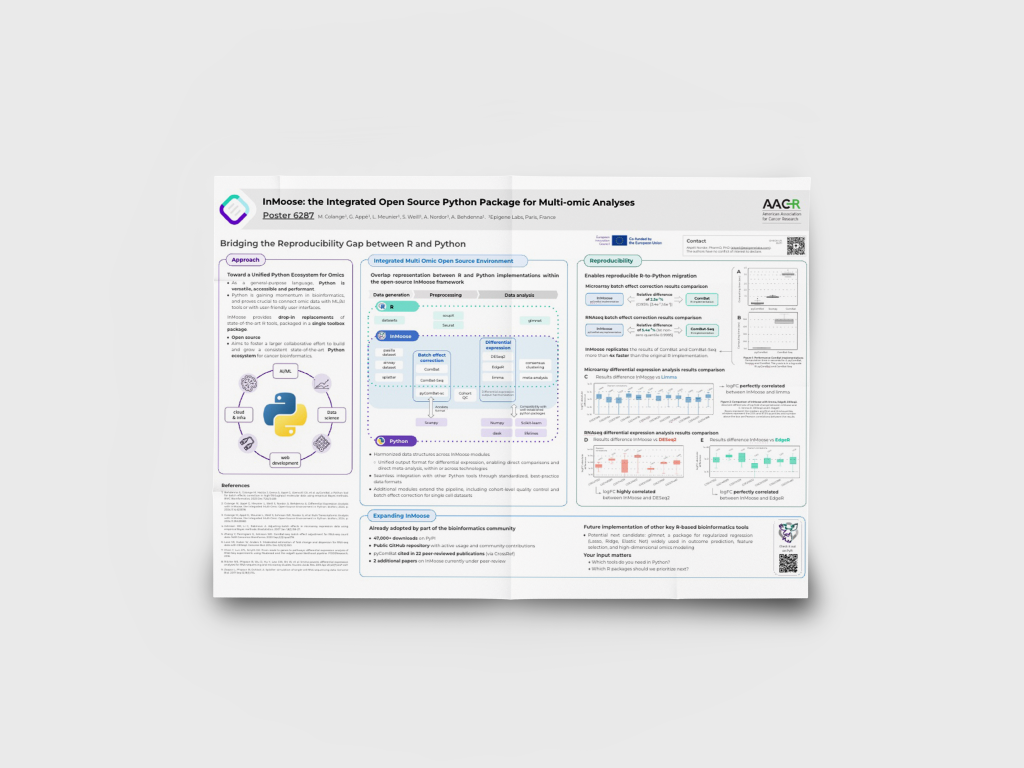
We developed a pan-cancer bioinformatics platform combining patient data with extensive biological and pharmaceutical knowledge.
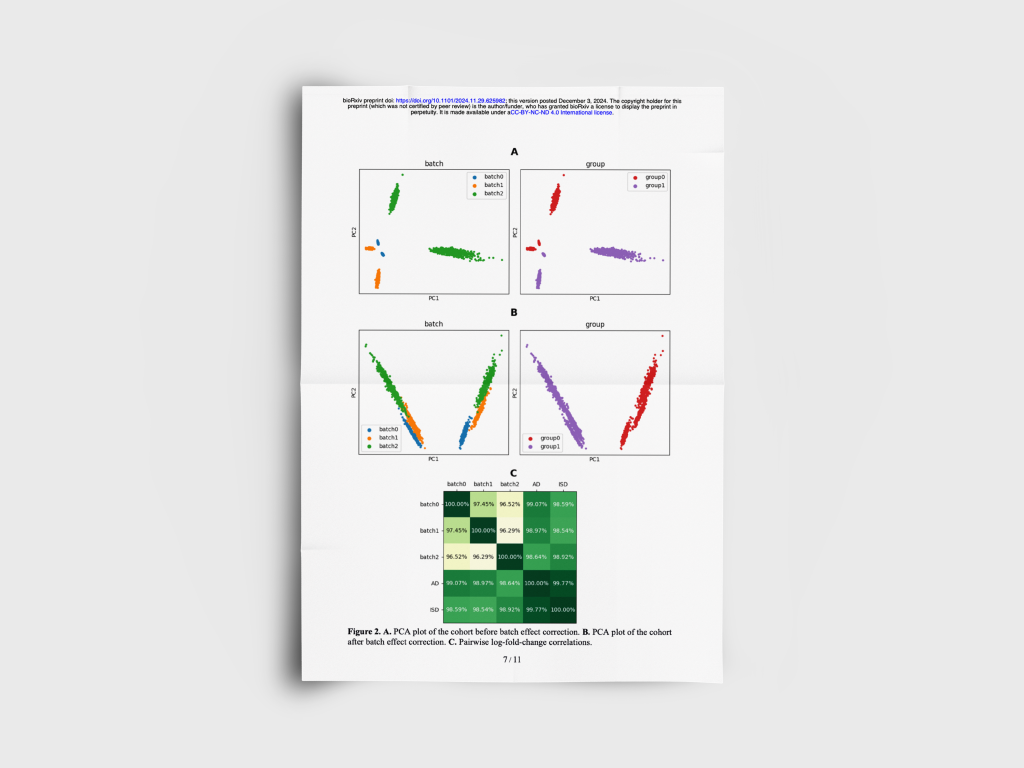
Empirical Bayesian batch effect correction for single-cell RNA-seq data

We developed a pan-cancer bioinformatics platform combining patient data with extensive biological and pharmaceutical knowledge.
Machine-learning-based inference of clinical metadata from gene expression data

We developed a pan-cancer bioinformatics platform combining patient data with extensive biological and pharmaceutical knowledge.
Bulk transcriptomic analysis with InMoose, the integrated multi-omic open-source environment in Python
We present InMoose, an open-source Python environment for omic data analysis, showcasing its capabilities in bulk transcriptomics.
Differential expression analysis with InMoose, the integrated multi-omic open-source environment in Python

We present the differential expression features of InMoose, a Python implementation of R tools limma, edgeR, and DESeq2.
Transforming public patient omic data into precision oncology targets

We present a scalable, data-driven platform for pan-cancer antigen target discovery leveraging the untapped potential of public transcriptomic data, along with extensive biological and pharmaceutical knowledge.
